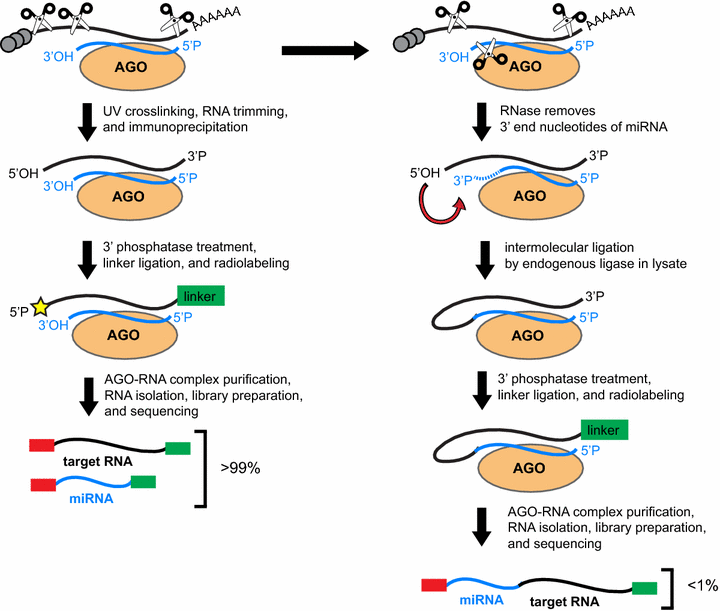
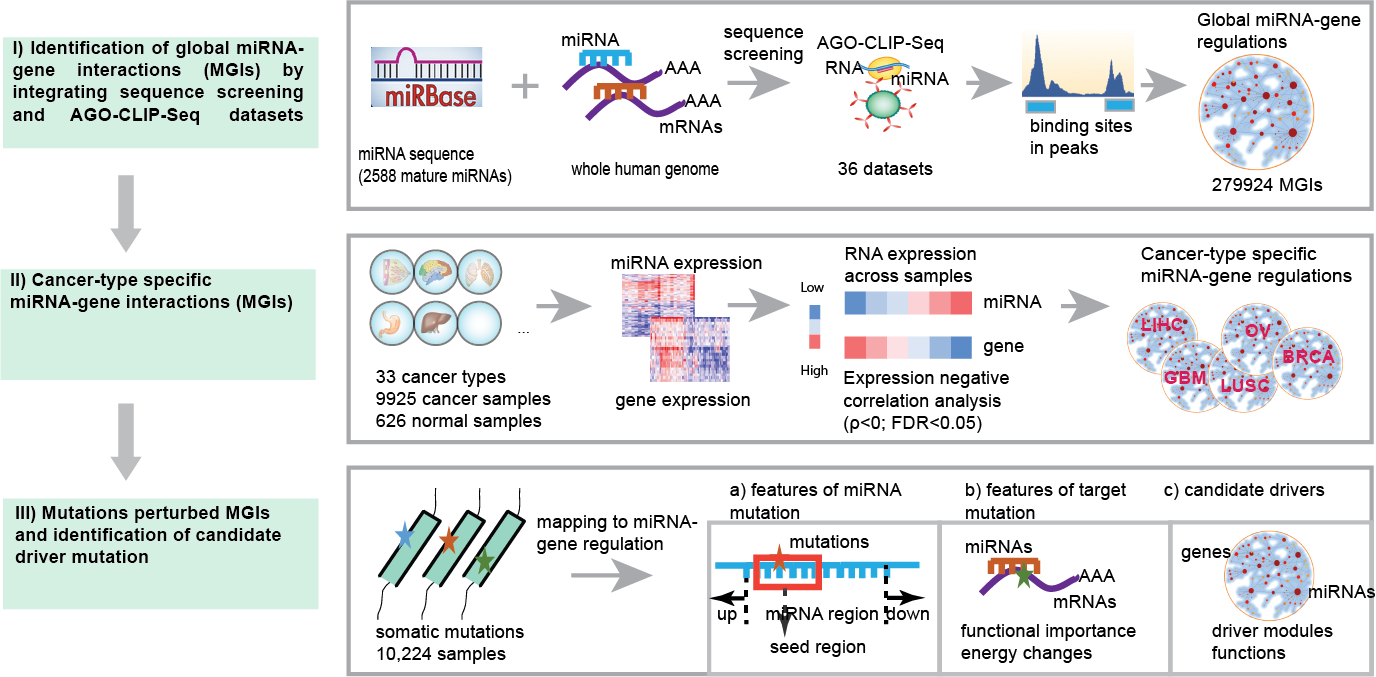
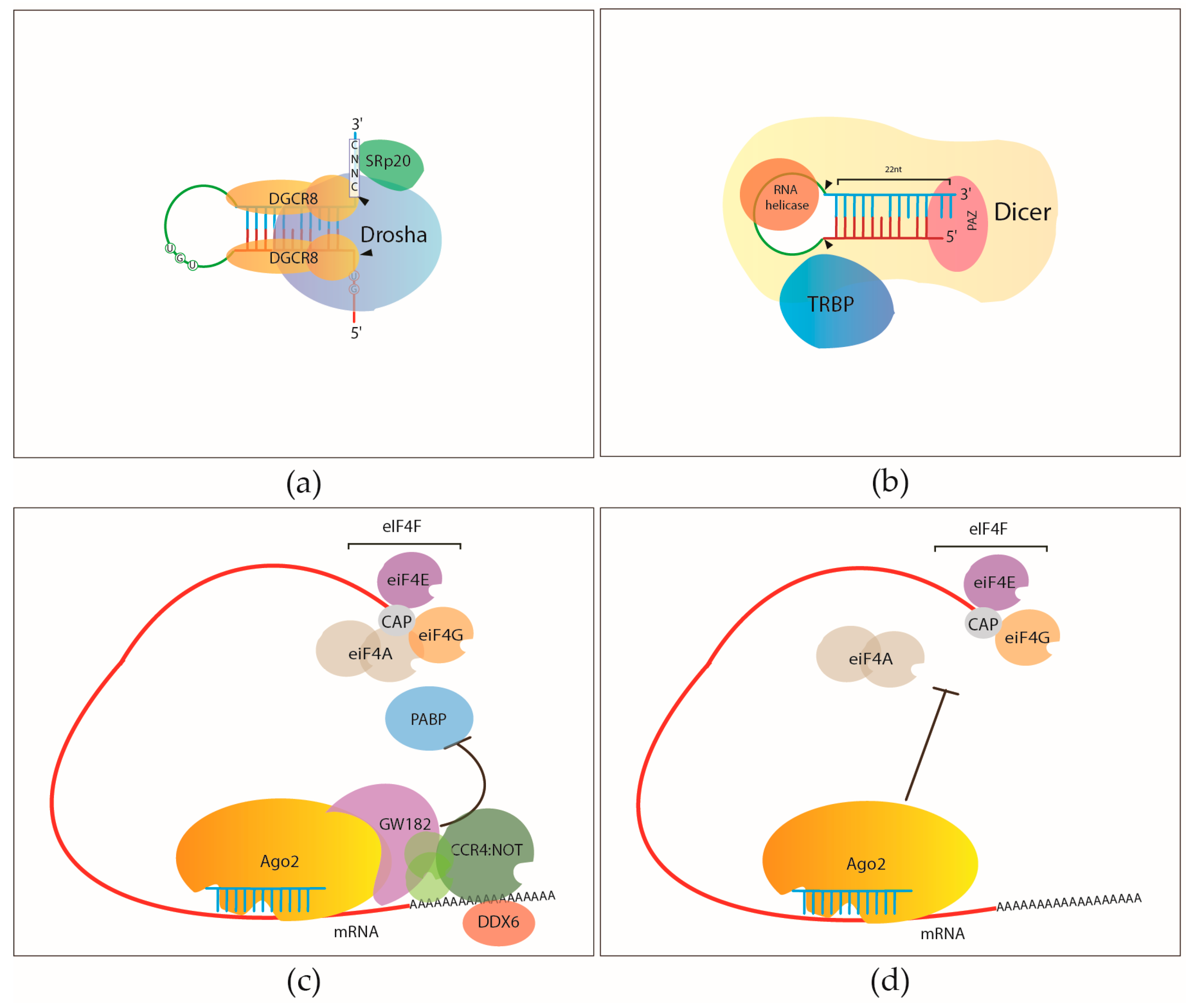
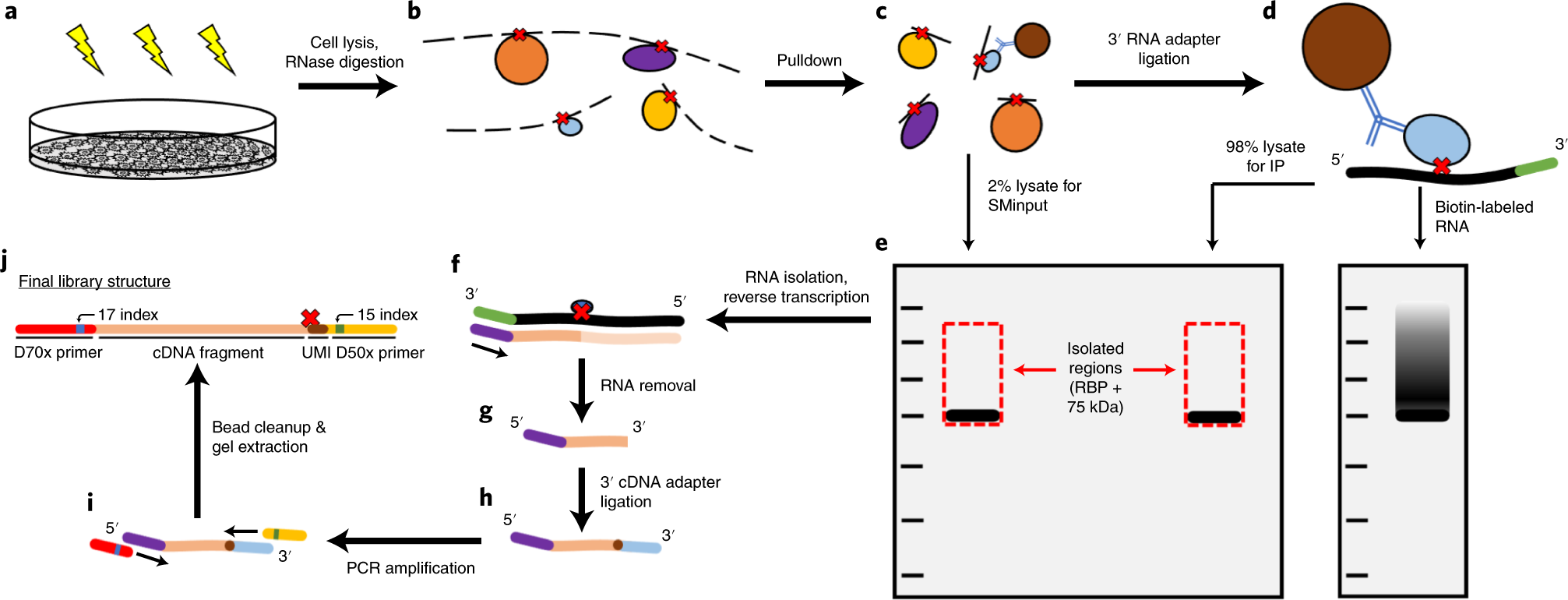
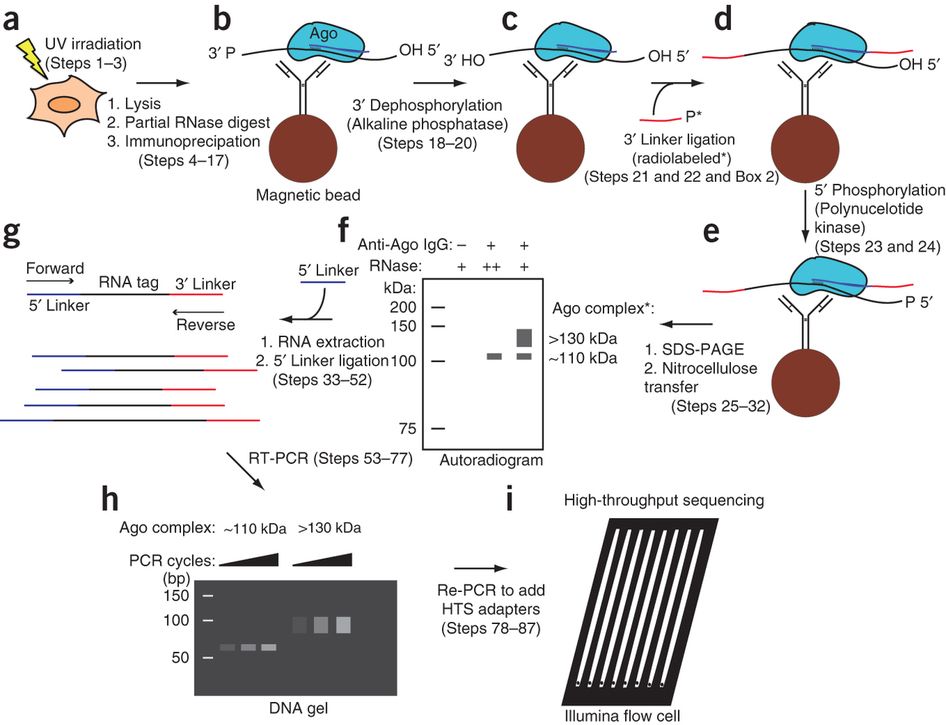
Interpretation of the AGO-binding model learned from CLIP-seq data. (A)... | Download Scientific Diagram

AGO CLIP Reveals an Activated Network for Acute Regulation of Brain Glutamate Homeostasis in Ischemic Stroke - ScienceDirect
AGO CLIP Reveals an Activated Network for Acute Regulation of Brain Glutamate Homeostasis in Ischemic Stroke

CLIP-seq overview. The CLIP-seq method for identifying miRNA target... | Download Scientific Diagram
![PDF] starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data | Semantic Scholar PDF] starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/265e0782aa831558f7dff55d820e880b1366fdf1/2-Figure1-1.png)
PDF] starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data | Semantic Scholar
Learning to Predict miRNA-mRNA Interactions from AGO CLIP Sequencing and CLASH Data | PLOS Computational Biology
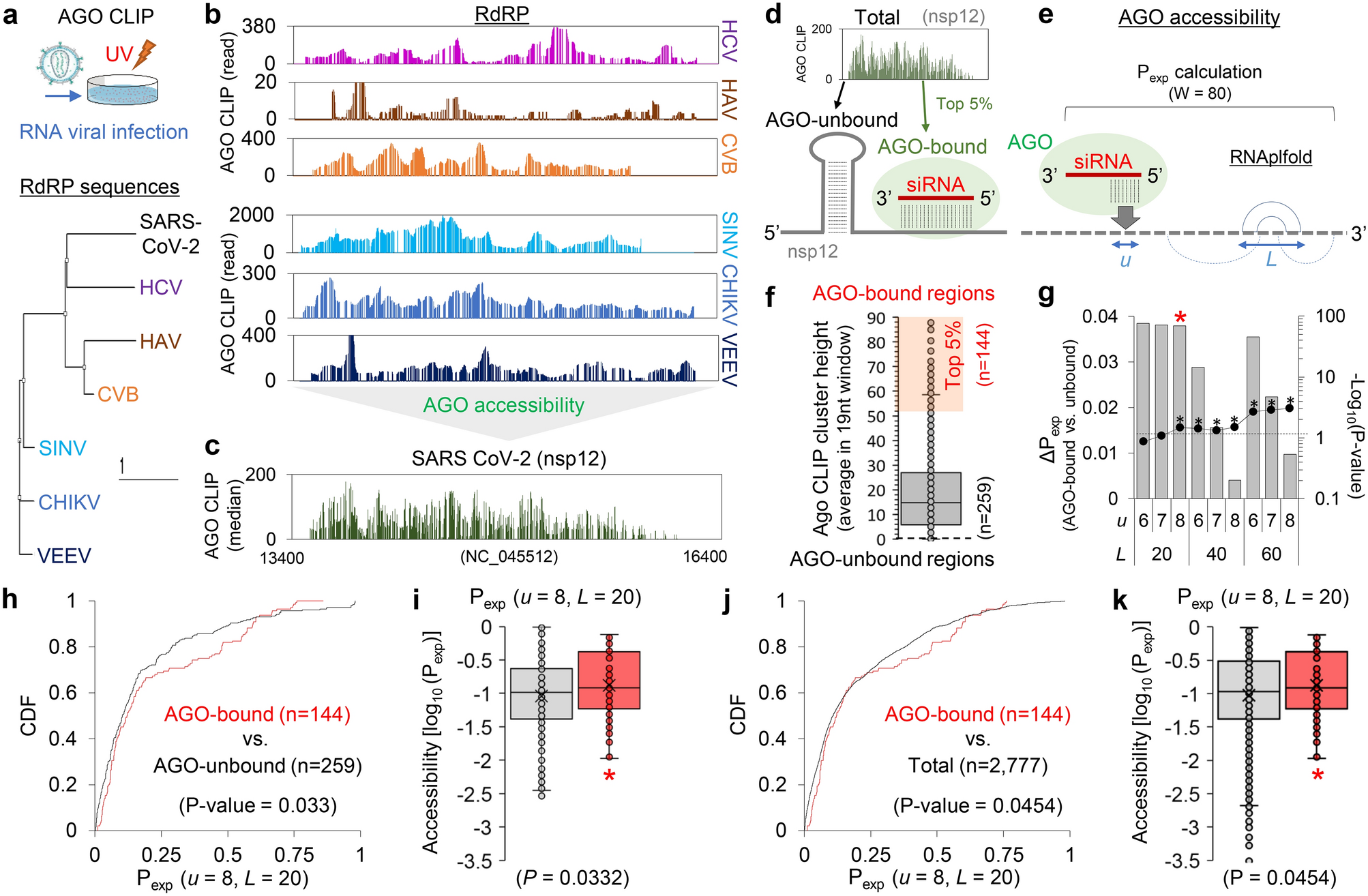
AGO CLIP-based imputation of potent siRNA sequences targeting SARS-CoV-2 with antifibrotic miRNA-like activity | Scientific Reports

High-Throughput Analysis Reveals miRNA Upregulating α-2,6-Sialic Acid through Direct miRNA–mRNA Interactions | ACS Central Science

circTools: identify and annotate circRNAs and their interactions with microRNAs (miRNAs) from large-scale <strong>CLIP-Seq (HITS-CLIP, PAR-CLIP, iCLIP, CLASH)</strong> and RNA-seq data.